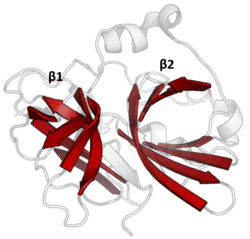
PA clan
| PA clan of proteases | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Identifiers | |||||||||
| Symbol | N/A | ||||||||
| Pfam | CL0124 | ||||||||
| InterPro | IPR009003 | ||||||||
| SCOP | 50494 | ||||||||
| SUPERFAMILY | 50494 | ||||||||
| |||||||||
The PA clan (Proteases of mixed nucleophile, superfamily A) is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but sequence identity of <10%. The clan contains both cysteine and serine proteases (different nucleophiles).[1][2] PA clan proteases can be found in plants,[3] animals,[3] fungi,[3] eubacteria,[4] archaea[5][6] and viruses.[2]
The PA clan represents an example of convergent evolution to the use of a catalytic triad for hydrolysis.[7] It also is an example of extreme divergent evolution of active sites in enzymes.[2]
History
In the 1960s, the sequence similarity of several proteases indicated that they were evolutionarily related.[8] These were grouped into the chymotrypsin-like serine proteases[9] (now called the S1 family). As the structures of these, and other proteases were solved by X-ray crystallography in the 1970s and 80s, it was noticed that several viral proteases such as Tobacco Etch Virus protease showed structural homology despite no discernible sequence similarity and even a different nucleophile.[2][10][11] Based on structural homology, a superfamily was defined and later named the PA clan (by the MEROPS classification system). As more structures are solved, more protease families have been added to the PA clan superfamily.[12][13]
Etymology
The P refers to Proteases of mixed nucleophile. The A indicates that it was the first such clan to be identified (there also exist the PB, PC, PD and PE clans).[1]
Structure
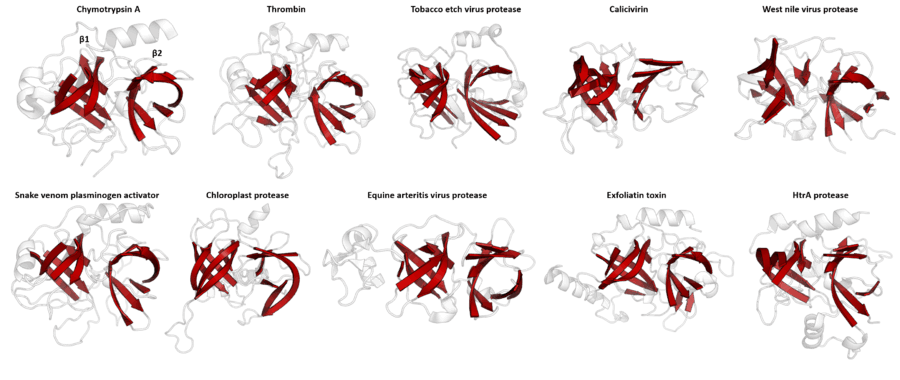

Despite retaining as little as 10% sequence identity, PA clan members isolated from viruses, prokaryotes and eukaryotes show structural homology and can be aligned by structural similarity (e.g. with DALI).[3]
Double β-barrel
PA clan proteases all share a core motif of two β-barrels with covalent catalysis performed by an acid-histidine-nucleophile catalytic triad motif. The barrels are arranged perpendicularly beside each other with hydrophobic residues holding them together as the core scaffold for the enzyme. The triad residues are split between the two barrels so that catalysis takes place at their interface.[14]
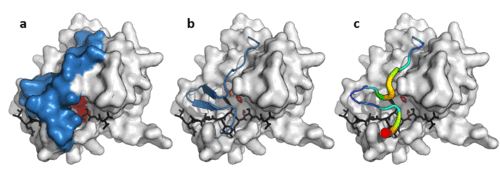
Viral protease loop
In addition to the double β-barrel core, some viral proteases (such as TEV protease) have a long, flexible C-terminal loop that forms a lid to which completely covers the substrate and create a binding tunnel. This tunnel contains a set of tight binding pockets such that each side chain of the substrate peptide (P6 to P1’) is bound in a complementary site (S6 to S1’) and specificity is endowed by the large contact area between enzyme and substrate.[11] Conversely, cellular proteases that lack this loop, such as trypsin have broader specificity.
Evolution and function
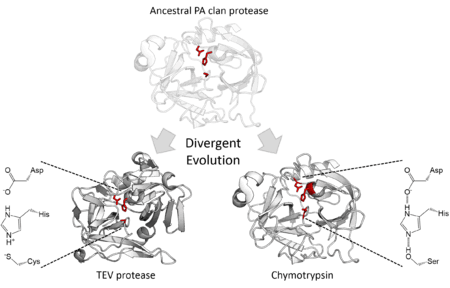
The structural homology described above indicates that the PA clan members are descended from a common ancestor of the same fold. Although PA clan proteases share a common ancestor some families use serine as the nucleophile in their catalytic triad whilst others use cysteine.[2] The catalytic triad is a set of three residues that together perform nucleophilic catalysis and are therefore a vital component of the enzyme active site. This is therefore an extreme example of divergent enzyme evolution since during evolutionary history, the core catalytic residue of the enzyme has switched.[15] In addition to their structural similarity, directed evolution has been shown to be able to convert a cysteine protease into an active serine protease.[16] All cellular PA clan proteases are serine proteases, however there are both serine and cysteine protease families of viral proteases.[7]
In addition to divergence in their core catalytic machinery, the PA clan proteases also show wide divergent evolution in function. Members of the PA clan can be found in eukaryotes, prokaryotes and viruses and encompass a wide range of functions. In mammals, some are involved in blood clotting (e.g. thrombin) and so have high substrate specificity as well as digestion (e.g. trypsin) with broad substrate specificity. Several snake venoms are also PA clan proteases, such as pit viper haemotoxin and interfere with the victim's blood clotting cascade. Additionally, bacteria such as Staphylococcus aureus secrete exfoliative toxin which digest and damage the host's tissues. Finally, many viruses express their genome as a single, massive polyprotein and use a protease to cleave this into functional units (e.g. polio, norovirus, and TEV proteases).[17][18]
Families
Within the PA clan (P=proteases of mixed nucleophiles), families are designated by their catalytic nucleophile (C=cysteine proteases, S=serine proteases). Despite the lack of sequence homology for the PA clan as a whole, individual families within it can be identified by sequence similarity.
| Family | Examples | Known structure? |
|---|---|---|
| C03 | Tobacco etch virus protease (tobacco etch virus ) | Yes |
| C04 | nuclear-inclusion-a peptidase (plum pox virus) | Yes |
| C24 | rabbit hemorrhagic disease virus 3C-like peptidase (rabbit hemorrhagic disease virus) | No |
| C30 | porcine transmissible gastroenteritis virus-type main peptidase (transmissible gastroenteritis virus) | Yes |
| C37 | calicivirin (Southampton virus) | Yes |
| C62 | gill-associated virus 3C-like peptidase (gill-associated virus) | No |
| C74 | pestivirus NS2 peptidase (bovine viral diarrhea virus 1) | No |
| C99 | iflavirus processing peptidase (Ectropis obliqua picorna-like virus) | No |
| S01 | chymotrypsin A (Bos taurus) | Yes |
| S03 | togavirin (Sindbis virus) | Yes |
| S06 | IgA specific serine endopeptidase (Neisseria gonorrhoeae) | Yes |
| S07 | flavivirin (yellow fever virus) | No |
| S29 | hepacivirin (hepatitis C virus) | Yes |
| S30 | potyvirus P1 peptidase (plum pox virus) | No |
| S31 | pestivirus NS3 polyprotein peptidase (bovine viral diarrhea virus 1) | No |
| S32 | equine arterivirus serine peptidase (equine arteritis virus) | Yes |
| S39 | sobemovirus peptidase (cocksfoot mottle virus) | Yes |
| S46 | dipeptidyl-peptidase 7 (Porphyromonas gingivalis) | No |
| S55 | SpoIVB peptidase (Bacillus subtilis) | No |
| S64 | Ssy5 peptidase (Saccharomyces cerevisiae) | No |
| S65 | picornain-like cysteine peptidase (Breda-1 torovirus) | No |
| S75 | White bream virus serine peptidase (White bream virus) | No |
See also
External resources
- MEROPS - Comprehensive protease database
- Superfamily - A database of protein folds
References
- 1 2 Rawlings ND, Barrett AJ, Bateman A (Jan 2012). "MEROPS: the database of proteolytic enzymes, their substrates and inhibitors". Nucleic Acids Research. 40 (Database issue): D343–50. doi:10.1093/nar/gkr987. PMC 3245014
 . PMID 22086950.
. PMID 22086950. - 1 2 3 4 5 Bazan JF, Fletterick RJ (Nov 1988). "Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications". Proceedings of the National Academy of Sciences of the United States of America. 85 (21): 7872–6. Bibcode:1988PNAS...85.7872B. doi:10.1073/pnas.85.21.7872. PMC 282299
 . PMID 3186696.
. PMID 3186696. - 1 2 3 4 Laskar A, Rodger EJ, Chatterjee A, Mandal C (May 24, 2012). "Modeling and structural analysis of PA clan serine proteases". BMC Research Notes. 5: 256. doi:10.1186/1756-0500-5-256. PMC 3434108
 . PMID 22624962.
. PMID 22624962. - ↑ Barbosa JA, Saldanha JW, Garratt RC (Jul 1996). "Novel features of serine protease active sites and specificity pockets: sequence analysis and modelling studies of glutamate-specific endopeptidases and epidermolytic toxins". Protein Engineering. 9 (7): 591–601. doi:10.1093/protein/9.7.591. PMID 8844831.
- ↑ "MEROPS - Archaeal S01 proteases". Retrieved 2013. Check date values in:
|access-date=(help) - ↑ Ruiz-Perez F, Nataro JP (Mar 2014). "Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence". Cellular and Molecular Life Sciences. 71 (5): 745–70. doi:10.1007/s00018-013-1355-8. PMID 23689588.
- 1 2 Buller AR, Townsend CA (Feb 2013). "Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad". Proceedings of the National Academy of Sciences of the United States of America. 110 (8): E653–61. Bibcode:2013PNAS..110E.653B. doi:10.1073/pnas.1221050110. PMC 3581919
 . PMID 23382230.
. PMID 23382230. - ↑ de Haën C, Neurath H, Teller DC (Feb 1975). "The phylogeny of trypsin-related serine proteases and their zymogens. New methods for the investigation of distant evolutionary relationships". Journal of Molecular Biology. 92 (2): 225–59. doi:10.1016/0022-2836(75)90225-9. PMID 1142424.
- ↑ Lesk AM, Fordham WD (May 1996). "Conservation and variability in the structures of serine proteinases of the chymotrypsin family". Journal of Molecular Biology. 258 (3): 501–37. doi:10.1006/jmbi.1996.0264. PMID 8642605.
- ↑ Gorbalenya AE, Blinov VM, Donchenko AP (Jan 1986). "Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families". FEBS Letters. 194 (2): 253–7. doi:10.1016/0014-5793(86)80095-3. PMID 3000829.
- 1 2 Phan J, Zdanov A, Evdokimov AG, Tropea JE, Peters HK, Kapust RB, Li M, Wlodawer A, Waugh DS (Dec 2002). "Structural basis for the substrate specificity of tobacco etch virus protease". The Journal of Biological Chemistry. 277 (52): 50564–72. doi:10.1074/jbc.M207224200. PMID 12377789.
- ↑ Allaire M, Chernaia MM, Malcolm BA, James MN (May 1994). "Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases". Nature. 369 (6475): 72–6. Bibcode:1994Natur.369...72A. doi:10.1038/369072a0. PMID 8164744.
- ↑ Snijder EJ, Wassenaar AL, van Dinten LC, Spaan WJ, Gorbalenya AE (Mar 1996). "The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases". The Journal of Biological Chemistry. 271 (9): 4864–71. doi:10.1074/jbc.271.9.4864. PMID 8617757.
- ↑ Dougherty WG, Parks TD, Cary SM, Bazan JF, Fletterick RJ (Sep 1989). "Characterization of the catalytic residues of the tobacco etch virus 49-kDa proteinase". Virology. 172 (1): 302–10. doi:10.1016/0042-6822(89)90132-3. PMID 2475971.
- ↑ Laskar, Aparna; Rodger, Euan J; Chatterjee, Aniruddha; Mandal, Chhabinath (2012-05-24). "Modeling and structural analysis of PA clan serine proteases". BMC Research Notes. 5 (1). doi:10.1186/1756-0500-5-256. ISSN 1756-0500. PMC 3434108
 . PMID 22624962.
. PMID 22624962. - ↑ Shafee, Thomas; Gatti-Lafranconi, Pietro; Minter, Ralph; Hollfelder, Florian (2015-09-07). "Handicap-Recover Evolution Leads to a Chemically Versatile, Nucleophile-Permissive Protease". ChemBioChem. 16 (13): 1866–1869. doi:10.1002/cbic.201500295. ISSN 1439-7633. PMC 4576821
 . PMID 26097079.
. PMID 26097079. - ↑ Salvesen, Guy (2013). Rawlings, Neil, ed. Handbook of proteolytic enzymes (3rd ed.). Boston: Academic Press. ISBN 9780123822192.
- ↑ Polgár, L. (2005-10-01). "The catalytic triad of serine peptidases". Cellular and molecular life sciences: CMLS. 62 (19-20): 2161–2172. doi:10.1007/s00018-005-5160-x. ISSN 1420-682X. PMID 16003488.