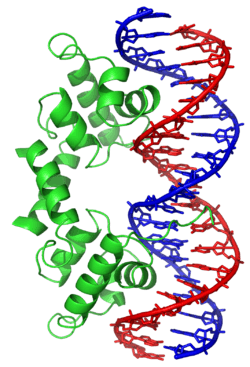
The
λ repressor of
bacteriophage lambda employs a helix-turn-helix (left; green) to bind
DNA (right; blue and red).
In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. It is composed of two α helices joined by a short strand of amino acids and is found in many proteins that regulate gene expression. It should not be confused with the helix-loop-helix domain.[1]
Discovery
The discovery of the helix-turn-helix motif was based on similarities between several genes encoding transcription regulatory proteins from bacteriophage lambda and Escherichia coli: Cro, CAP, and λ repressor, which were found to share a common 20-25 amino acid sequence that facilitates DNA recognition.[2][3][4][5]
Function
The helix-turn-helix motif is a DNA-binding motif. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminal end of the motif, the other at the C-terminus. In most cases, such as in the Cro repressor, the second helix contributes most to DNA recognition, and hence it is often called the "recognition helix". It binds to the major groove of DNA through a series of hydrogen bonds and various Van der Waals interactions with exposed bases. The other α helix stabilizes the interaction between protein and DNA, but does not play a particularly strong role in its recognition.[2] The recognition helix and its preceding helix always have the same relative orientation.[6]
Classification of helix-turn-helix motifs
Several attempts have been made to classify the helix-turn-helix motifs based on their structure and the spatial arrangement of their helices.[6][7][8] Some of the main types are described below.
Di-helical
The di-helical helix-turn-helix motif is the simplest helix-turn-helix motif. A fragment of Engrailed homeodomain encompassing only the two helices and the turn was found to be an ultrafast independently folding protein domain.[9]
Tri-helical
An example of this motif is found in the Transcriptional activator Myb.[10]
Tetra-helical
The tetra-helical helix-turn-helix motif has an additional C-terminal helix compared to the tri-helical motifs. These include the LuxR-type DNA-binding HTH domain found in bacterial transcription factors and the helix-turn-helix motif found in the TetR repressors.[11] Multihelical versions with additional helices also occur.[12]
Winged helix-turn-helix
The winged helix-turn-helix (wHTH) motif is formed by a 3-helical bundle and a 3- or 4-strand beta-sheet (wing). The topology of helices and strands in the wHTH motifs may vary. In the transcription factor ETS wHTH folds into a helix-turn-helix motif on a four-stranded anti-parallel beta-sheet scaffold arranged in the order α1-β1-β2-α2-α3-β3-β4 where the third helix is the DNA recognition helix.[13][14]
Other modified helix-turn-helix motifs
Other derivatives of the helix-turn-helix motif include the DNA-binding domain found in MarR, a regulator of multiple antibiotic resistance, which forms a winged helix-turn-helix with an additional C-terminal alpha helix.[8][15]
See also
References
- ↑ Brennan RG, Matthews BW (1989). "The helix-turn-helix DNA binding motif.". J Biol Chem. 264 (4): 1903–6. PMID 2644244.
- 1 2 Matthews BW, Ohlendorf DH, Anderson WF, Takeda Y (1982). "Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor.". Proc Natl Acad Sci U S A. 79 (5): 1428–32. doi:10.1073/pnas.79.5.1428. PMC 345986
 . PMID 6951187.
. PMID 6951187. - ↑ Anderson WF, Ohlendorf DH, Takeda Y, Matthews BW (1981). "Structure of the cro repressor from bacteriophage lambda and its interaction with DNA.". Nature. 290 (5809): 754–8. doi:10.1038/290754a0. PMID 6452580.
- ↑ McKay DB, Steitz TA (1981). "Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA.". Nature. 290 (5809): 744–9. doi:10.1038/290744a0. PMID 6261152.
- ↑ Pabo CO, Lewis M (1982). "The operator-binding domain of lambda repressor: structure and DNA recognition.". Nature. 298 (5873): 443–7. doi:10.1038/298443a0. PMID 7088190.
- 1 2 Wintjens R, Rooman M (1996). "Structural classification of HTH DNA-binding domains and protein-DNA interaction modes.". J Mol Biol. 262 (2): 294–313. doi:10.1006/jmbi.1996.0514. PMID 8831795.
- ↑ Suzuki M, Brenner SE (1995). "Classification of multi-helical DNA-binding domains and application to predict the DBD structures of sigma factor, LysR, OmpR/PhoB, CENP-B, Rapl, and Xy1S/Ada/AraC.". FEBS Lett. 372 (2-3): 215–21. doi:10.1016/0014-5793(95)00988-L. PMID 7556672.
- 1 2 Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005). "The many faces of the helix-turn-helix domain: transcription regulation and beyond.". FEMS Microbiol Rev. 29 (2): 231–62. doi:10.1016/j.femsre.2004.12.008. PMID 15808743.
- ↑ Religa TL, Johnson CM, Vu DM, Brewer SH, Dyer RB, Fersht AR (2007). "The helix-turn-helix motif as an ultrafast independently folding domain: the pathway of folding of Engrailed homeodomain.". Proc Natl Acad Sci U S A. 104 (22): 9272–9277. doi:10.1073/pnas.0703434104. PMID 17517666.
- ↑ Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, et al. (1992). "Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core.". Proc Natl Acad Sci U S A. 89 (14): 6428–32. doi:10.1073/pnas.89.14.6428. PMC 49514
 . PMID 1631139.
. PMID 1631139. - ↑ Hinrichs W, Kisker C, Düvel M, Müller A, Tovar K, Hillen W, et al. (1994). "Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance.". Science. 264 (5157): 418–20. doi:10.1126/science.8153629. PMID 8153629.
- ↑ Iwahara J, Clubb RT (1999). "Solution structure of the DNA binding domain from Dead ringer, a sequence-specific AT-rich interaction domain (ARID).". EMBO J. 18 (21): 6084–94. doi:10.1093/emboj/18.21.6084. PMC 1171673
 . PMID 10545119.
. PMID 10545119. - ↑ Donaldson LW, Petersen JM, Graves BJ, McIntosh LP (1996). "Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif". EMBO J. 15 (1): 125–34. doi:10.2210/pdb1etc/pdb. PMC 449924
 . PMID 8598195.
. PMID 8598195. - ↑ Sharrocks AD, Brown AL, Ling Y, Yates PR (1997). "The ETS-domain transcription factor family". Int. J. Biochem. Cell Biol. 29 (12): 1371–87. doi:10.1016/S1357-2725(97)00086-1. PMID 9570133.
- ↑ Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF (2001). "The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution.". Nat Struct Biol. 8 (8): 710–4. doi:10.1038/90429. PMID 11473263.
Further reading
External links
| Pfam infoboxes for Helix-turn-helix domains |
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| putative Helix-turn-helix domain of transposase IS66 |
|---|
| Identifiers |
|---|
| Symbol |
HTH_Tnp_IS66 |
|---|
| Pfam |
PF13005 |
|---|
| Pfam clan |
CL0123 |
|---|
|
|
|
|
|
|
|---|
|
|
|
(2) Zinc finger DNA-binding domains |
|---|
| | (2.1) Nuclear receptor (Cys4) | | subfamily 1 | |
|---|
| | subfamily 2 | |
|---|
| | subfamily 3 | |
|---|
| | subfamily 4 | |
|---|
| | subfamily 5 | |
|---|
| | subfamily 6 | |
|---|
| | subfamily 0 | |
|---|
|
|---|
| | (2.2) Other Cys4 | |
|---|
| | (2.3) Cys2His2 | |
|---|
| | (2.4) Cys6 | |
|---|
| | (2.5) Alternating composition | |
|---|
| | (2.6) WRKY | |
|---|
|
|
|
|
|
| (4) β-Scaffold factors with minor groove contacts |
|---|
| |
|
|
| (0) Other transcription factors |
|---|
| |
|
|
see also transcription factor/coregulator deficiencies |

 . PMID 6951187.
. PMID 6951187. . PMID 1631139.
. PMID 1631139. . PMID 10545119.
. PMID 10545119. . PMID 8598195.
. PMID 8598195.