EcoRI
| EcoRI | |||||||||
|---|---|---|---|---|---|---|---|---|---|
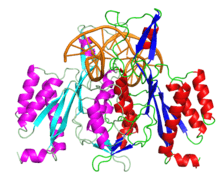
 The crystallographic structure of restriction endonuclease EcoRI at 3.3 a in the absence of DNA | |||||||||
| Identifiers | |||||||||
| Symbol | EcoRI | ||||||||
| Pfam | PF02963 | ||||||||
| InterPro | IPR004221 | ||||||||
| SCOP | 1na6 | ||||||||
| SUPERFAMILY | 1na6 | ||||||||
| |||||||||
EcoRI (pronounced, "eco R one") is a restriction endonuclease enzyme isolated from species E. coli. The Eco part of the enzyme's name originates from the species from which it was isolated, while the R represents the particular strain, in this case RY13. The last part of its name, the I, denotes that it was the first enzyme isolated from this strain. EcoRI is a restriction enzyme that cleaves DNA double helixes into fragments at specific sites. It is also a part of the restriction modification system.
In molecular biology it is used as a restriction enzyme. EcoRI creates 5 nucleotide sticky ends with 5' end overhangs of AATTC. The nucleic acid recognition sequence where the enzyme cuts is G/AATTC, which has a palindromic, complementary sequence of CTTAA/G. The / in the sequence indicates which phosphodiester bond the enzyme will break in the DNA molecule. Other restriction enzymes, depending on their cut sites, can also leave 3' overhangs or blunt ends with no overhangs.
Structure
Primary structure
EcoRI contains the PD..D/EXK motif within its active site like many restriction endonucleases.
Tertiary and quaternary structure
The enzyme is a homodimer of a 31 kilodalton subunit consisting of one globular domain of the α/β architecture. Each subunit contains a loop which sticks out from the globular domain and wraps around the DNA when bound.[1][2]

EcoRI has been cocrystallized with the sequence it normally cuts. This crystal was used to solve the structure of the complex 1QPS. The solved crystal structure shows that the subunits of the enzyme homodimer interact with the DNA symmetrically.[1] In the complex, two α-helices from each subunit come together to form a four-helix bundle.[3] On the interacting helices are residues Glu144 and Arg145, which interact together, forming a crosstalk ring that is believed to allow the enzyme's two active sites to communicate.[4]
Uses
Restriction enzymes, such as EcoRI, are used in a wide variety of molecular genetics techniques including cloning, DNA screening and deleting sections of DNA in vitro. Restriction enzymes, like EcoRI, that generate sticky ends of DNA are often used to cut DNA prior to ligation, as the sticky ends make the ligation reaction more efficient. EcoRI can exhibit non-site-specific cutting, known as star activity, depending on the conditions present in the reaction. Conditions that can induce star activity when using EcoRI include low salt concentration, high glycerol concentration, excessive amounts of enzyme present in the reaction, high pH and contamination with certain organic solvents.[5]
See also
References
- 1 2 Pingoud, A.; Jeltsch, A. (2001). "Structure function of type II restriction endonucleases". Nucl. Acids Res. 29 (18): 3705–3727. doi:10.1093/nar/29.18.3705. PMC 55916
 . PMID 11557805.
. PMID 11557805. - ↑ Kurpiewski, M. R.; Engler, L. E.; Wozniak, L. A.; Kobylanska, A.; Koziolkiewicz, M.; Stec, W. J; Jen-Jacobson, L (2004). "Mechanisms of coupling between DNA recognition catalysis in EcoRI endonucleases". Structure. 12: 1775–1788. doi:10.1016/j.str.2004.07.016. PMID 15458627.
- ↑ Bitinaite, J. D. A.; Wah Aggarwal, A. K.; Schildkraut, I. (1998). "FokI dimerization is required for DNA cleavage". Proc Natl Acad Sci U S A. 95 (18): 10570–5. doi:10.1073/pnas.95.18.10570. PMC 27935
 . PMID 9724744.
. PMID 9724744. - ↑ Kim, Y. C.; Grable, J. C.; Love, R.; Greene, P. J.; Rosenberg, J. M. (1990). "Refinement of EcoRI endonuclease crystal structure: a revised protein chain tracing". Science. 249 (4974): 1307–1309. doi:10.1126/science.2399465. PMID 2399465.
- ↑ http://www.neb.com/nebecomm/products/faqproductR0101.asp#1
External links
- Eco-RI at the US National Library of Medicine Medical Subject Headings (MeSH)