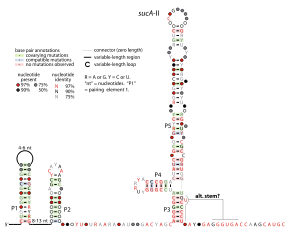
SucA-II RNA motif
| sucA-II RNA | |
|---|---|
|
Consensus secondary structure of sucA-II RNAs | |
| Identifiers | |
| Symbol | sucA-II RNA |
| Rfam | RF01758 |
| Other data | |
| RNA type | Cis-regulatory element |
| Domain(s) | Pseudomonas |
The sucA-II RNA motif is a conserved RNA structure identified by bioinformatics.[1] It is consistently found in the presumed 5' untranslated regions of sucA genes, which encode Oxoglutarate dehydrogenase enzymes that participate in the citric acid cycle. Given this arrangement, sucA-II RNAs might regulate the downstream sucA gene. This genetic arrangement is similar to the previously reported sucA RNA motif. However, sucA-II RNAs are found only in bacteria classified within the genus Pseudomonas, whereas the previously reported motif is found only in betaproteobacteria.
See also
References
- ↑ Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol. 11 (3): R31. doi:10.1186/gb-2010-11-3-r31. PMC 2864571
 . PMID 20230605.
. PMID 20230605.
External links
This article is issued from Wikipedia - version of the 9/2/2015. The text is available under the Creative Commons Attribution/Share Alike but additional terms may apply for the media files.