R-loop
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA (ssDNA). R-loops may be formed in a variety of circumstances, and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety.
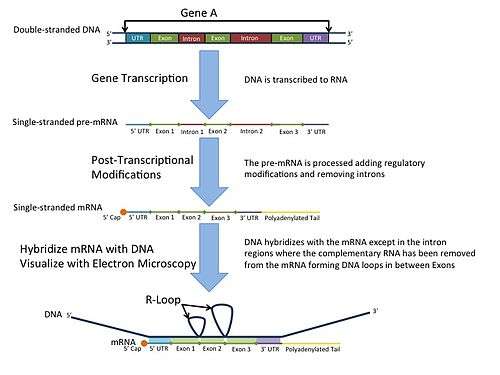
In the laboratory, R-loops may also be created by the hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been spliced out of the mRNA) form single-stranded loops, as they cannot hybridize with complementary sequence in the mRNA.
History
R-looping was first described in 1976.[1] Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that protein coding adenovirus genes contained DNA sequences that were not present in the mature mRNA.[2][3] Roberts and Sharp were awarded the Nobel Prize in 1993 for independently discovering introns. After their discovery in adenovirus, introns were found in a number of eukaryotic genes such as the eukaryotic ovalbumin gene (first by the O'Malley laboratory, then confirmed by other groups) [4][5] hexon DNA,[6] and extrachromosomal rRNA genes of Tetrahymena thermophila.[7]
In the mid-1980s, development of an antibody that binds specifically to the R-loop structure opened the door for immunofluorescence studies, as well as genome-wide characterization of R-loop formation by DRIP-seq.[8]
R-loop mapping
R-loop mapping is a laboratory technique used to distinguish introns from exons in double-stranded DNA.[9] These R-loops are visualized by electron microscopy and reveal intron regions of DNA by creating unbound loops at these regions.[10]
R-loops in vivo
The potential for R-loops to serve as replication primers was demonstrated in 1980.[11] In 1994, R-loops were demonstrated to be present in vivo through analysis of plasmids isolated from E. coli mutants carrying mutations in topoisomerase.[12] This discovery of endogenous R-loops, in conjunction with rapid advances in genetic sequencing technologies, inspired a blossoming of R-loop research in the early 2000s that continues to this day.[13]
Regulation of R-loop formation and resolution
RNaseH enzymes are the primary proteins responsible for the dissolution of R-loops, acting to degrade the RNA moiety in order to allow the two complementary DNA strands to anneal.[14] Research over the past decade has identified more than 50 proteins that appear to influence R-loop accumulation, and while many of them are believed to contribute by sequestering or processing newly transcribed RNA to prevent re-annealing to the template, mechanisms of R-loop interaction for many of these proteins remain to be determined.[15]
Roles of R-loops in genetic regulation
R-loop formation is a key step in immunoglobulin class switching, a process that allows activated B cells to modulate antibody production.[16] They also appear to play a role in protecting some active promoters from methylation.[17] Additionally, R-loop formation appears to be associated with “open” chromatin, characteristic of actively transcribed regions.[18][19]
R-loops as genetic damage
When unscheduled R-loops form, they can cause damage by a number of different mechanisms. Exposed ssDNA can come under attack by endogenous mutagens, including DNA-modifying enzymes such as Activation-induced cytidine deaminase, and can block replication forks to induce fork collapse and subsequent double strand breaks (DSBs).[20] As well, R-loops may induce unscheduled replication by acting as a primer.[21][22]
R-loop accumulation has been associated with a number of diseases, including amyotrophic lateral sclerosis type 4 (ALS4), ataxia oculomotor apraxia type 2 (AOA2), Aicardi–Goutières syndrome, Angelman syndrome, Prader–Willi syndrome, and cancer.[23]
See also
References
- ↑ Thomas, M; White, RL; Davis, RW (1976). "Hybridization of RNA to double-stranded DNA: formation of R-loops". Proc. Natl. Acad. Sci. USA. 73: 2294–2298. doi:10.1073/pnas.73.7.2294.
- ↑ Berget, SM; Moore, C; Sharp, PA (1977). "Spliced segments at the 5′ terminus of adenovirus 2 late mRNA". Proc Natl Acad Sci USA. 8: 3171–3175. doi:10.1073/pnas.74.8.3171. PMC 431482
 . PMID 269380.
. PMID 269380. - ↑ Chow, LT; Gelinas, RE; Broker, TR; Roberts, RJ (1977). "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA". Cell. 12 (1): 1–8. doi:10.1016/0092-8674(77)90180-5. PMID 902310.
- ↑ Lai, EC; Woo, SL; Dugaiczyk, A; Catterall, JF; O'Malley, BW (May 1978). "The ovalbumin gene: structural sequences in native chicken DNA are not contiguous". Proc Natl Acad Sci U S A. 75 (5): 2205–9. doi:10.1073/pnas.75.5.2205.
- ↑ O'Hare, K; Breathnach, R; Benoist, C; Chambon, P (1979). "No more than seven interruptions in the ovalbumin gene: comparison of genomic and double-stranded cDNA sequences". Oxford Journals. 7 (2): 321–334. doi:10.1093/nar/7.2.321.
- ↑ Berget, SM; Moore, C; Sharp, PA (1977). "Spliced segments at the 5′ terminus of adenovirus 2 late mRNA". Proc Natl Acad Sci USA. 8: 3171–3175. doi:10.1073/pnas.74.8.3171. PMC 431482
 . PMID 269380.
. PMID 269380. - ↑ Cech, TR; Rio, DC (1979). "Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping". Proc Natl Acad Sci USA. 76 (10): 5051–5055. doi:10.1073/pnas.76.10.5051.
- ↑ Boguslawski, SJ; et al. (1986). "Characterization of monoclonal antibody to DNA. RNA and its application to immunodetection of hybrids". J Immunol Methods. 89 (1): 123–30. doi:10.1016/0022-1759(86)90040-2.
- ↑ Woolford, John L.; Jr; Rosbash, Michael (1979). "The use of R-looping for structural gene identification and mRNA purification". Nucleic Acids Research. 6 (7): 2483–97. doi:10.1093/nar/6.7.2483.
- ↑ King RC, Stansfield WD, Mulligan PK (2007). A Dictionary of Genetics. Oxford University Press 7.
- ↑ Itoh, T; Tomizawa, J (1980). "Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H.". Proc Natl Acad Sci USA. 77: 2450–2454. doi:10.1073/pnas.77.5.2450.
- ↑ Drolet, M; Bi, X; Liu, LF (1994). "Hypernegative supercoiling of the DNA template during transcription elongation in vitro". J Biol Chem. 269: 2068–2074.
- ↑ Groh, M; Gromak, N (2014). "Out of Balance: R-loops in Human Disease". PLoS Genet. 10 (9): e1004630. doi:10.1371/journal.pgen.1004630.
- ↑ Cerritelli, SM; Crouch, RJ (2009). "Ribonuclease H: the enzymes in eukaryotes". FEBS J. 279: 1494–1505.
- ↑ Chan, YA; Aristizabal, MJ; Lu, PY; Luo, Z; Hamza, A; Kobor, MS; Stirling, PC; Hieter, P (2014). "Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip". PLOS Genet. 10: e1004288. doi:10.1371/journal.pgen.1004288. PMC 3990523
 . PMID 24743342.
. PMID 24743342. - ↑ Roy, D; Yu, K; Lieber, MR (2008). "Mechanism of R-loop formation at immunoglobulin class switch sequences". Mol Cell Biol. 28: 50–60. doi:10.1128/mcb.01251-07.
- ↑ Ginno, PA; Lott, PL; Christensen, HC; Korf, I; Chedin, F (2012). "R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters". Mol Cell. 45: 814–825. doi:10.1016/j.molcel.2012.01.017. PMID 22387027.
- ↑ Castellano-Pozo, M; Santos-Pereira, JM; Rondon, AG; Barroso, S; Andujar, E; Perez-Alegre, M; Garcia-Muse, T; Aguilera, A (2013). "R loops are linked to histone H3 S10 phosphorylation and chromatin condensation". Mol Cell. 52: 583–590. doi:10.1016/j.molcel.2013.10.006. PMID 24211264.
- ↑ Costantino, L; Koshland, D (2015). "The Yin and Yang of R-loop biology". Curr Opin Cell Biol. 34: 39–45. doi:10.1016/j.ceb.2015.04.008.
- ↑ Sollier, J; Cimprich, KA (2015). "Breaking bad: R-loops and genome integrity". Trends Cell Biol. 25: 514–522. doi:10.1016/j.tcb.2015.05.003.
- ↑ Itoh, T; Tomizawa, J (1980). "Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H.". Proc Natl Acad Sci USA. 77: 2450–2454. doi:10.1073/pnas.77.5.2450.
- ↑ Costantino, L; Koshland, D (2015). "The Yin and Yang of R-loop biology". Curr Opin Cell Biol. 34: 39–45. doi:10.1016/j.ceb.2015.04.008.
- ↑ Groh, M; Gromak, N (2014). "Out of Balance: R-loops in Human Disease". PLoS Genet. 10 (9): e1004630. doi:10.1371/journal.pgen.1004630.