NADH peroxidase
| NADH peroxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Identifiers | |||||||||
| EC number | 1.11.1.1 | ||||||||
| CAS number | 9032-24-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In enzymology, a NADH peroxidase (EC 1.11.1.1) is an enzyme that catalyzes the chemical reaction
- NADH + H+ + H2O2 NAD+ + 2 H2O
The presumed function of NADH peroxidase is to inactivate H2O2 generated within the cell, for example by glycerol-3-phosphate oxidase during glycerol metabolism or dismutation of superoxide, before the H2O2 causes damage to essential cellular components.[1]
The 3 substrates of this enzyme are NADH, H+, and H2O2, whereas its two products are NAD+ and H2O. It employs one cofactor, FAD, however no discrete FADH2 intermediate has been observed.[2]
This enzyme belongs to the family of oxidoreductases, specifically those acting on a peroxide as acceptor (peroxidases). The systematic name of this enzyme class is NADH:hydrogen-peroxide oxidoreductase. Other names in common use include DPNH peroxidase, NAD peroxidase, diphosphopyridine nucleotide peroxidase, NADH-peroxidase, nicotinamide adenine dinucleotide peroxidase, and NADH2 peroxidase.
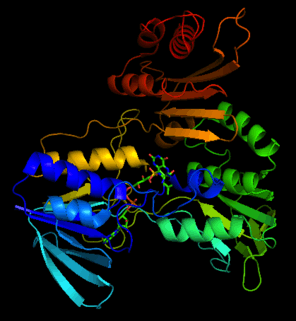
Structure
The crystal structure of NADH peroxidase resembles glutathione reductase with respect to chain fold and location as well as conformation of the prosthetic group FAD[3]
His10 of the NADH peroxidase is located near the N-terminus of the R1 helix within the FAD-binding site.[4] One of the oxygen atoms of Cys42-SO3H is hydrogen-bonded both to the His10 imidazole and to Cys42 N terminus. The His10 functions in part to stabilize the unusual Cys42-SOH redox center.[3] Arg303 also stabilizes the Cys42-SO3H. Glu-14 participates in forming the tight dimer interface that limits solvent accessibility, important for maintaining the oxidation state of the sulfenic acid.[4]
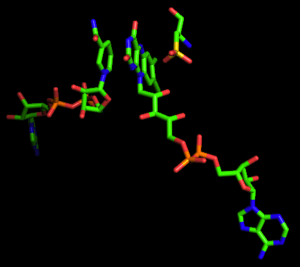
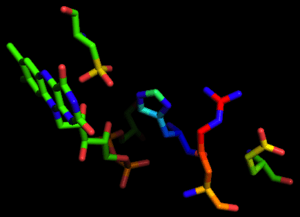
Reaction mechanism
The NADH peroxidase from Enterococcus faecalis is unique in that it utilizes the Cys42 thiol/sulfenic acid (-SH/-SOH) redox couple in the heterolytic cleavage of the peroxide bond to catalyze the two-electron reduction of hydrogen peroxide to water.[5]
The kinetic mechanism of the wild-type peroxidase involves (1) NADH reduction of E(FAD, Cys42-SOH) to EH2(FAD, Cys42-SH) in an initial priming step; (2) rapid binding of NADH to EH2; (3) reduction of H2O2 by the Cys42-thiolate, yielding E•NADH; and (4) rate-limiting hydride transfer from bound NADH, regenerating EH2.[6] No discrete FADH2 intermediate has been observed, however, and the precise details of Cys42-SOH reduction have not been elucidated.[7]
- E + NADH → (EH2'•NAD+)* → EH2'•NAD+ → EH2 + NAD+ + H2O
- EH2 + NADH → EH2•NADH*
- EH2•NADH* + H2O2 → E•NADH + H2O
- E•NADH + H+ → EH2•NAD+ + H2O
- EH2•NAD+ → EH2 + NAD+
Inhibitors include Ag+, Cl−, Co2+, Cu2+, Hg2+, NaN3, Pb2+, and SO42−.[8] At suboptimal H2O2 concentrations and concentrations of NADH that are saturating, NADH inhibits the peroxidase activity of the NADH peroxidase by converting the enzyme to an unstable intermediate. NAD+ behaves as an activator by reversing the equilibria that lead to the unstable intermediate, thus converting the enzyme to the kinetically active complex that reduces H2O2.[9]
Biological Function
NADH eliminates potentially toxic hydrogen peroxide under aerobic growth conditions and represents an enzymatic defense available against H2O2-mediated oxidative stress. Second, the enzyme presents an additional mechanism for regeneration of the NAD+ essential to the strictly fermentative metabolism of this organism.[2][10] The enzyme may also protect against exogenous H2O2 and contribute to bacterial virulence.[11]
The actual function of NADH peroxidases and oxidases in plants is still unclear, but they could act in early signaling of oxidative stress through producing H2O2.[12]
An alternative role may include regulation of H2O2 formation by NADH peroxidase and oxidase in cell wall loosening and reconstruction.[13]
References
- ↑ La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A (December 2007). "Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis". Mol. Microbiol. 66 (5): 1148–63. doi:10.1111/j.1365-2958.2007.05987.x. PMID 17971082.
- 1 2 Miller H, Poole LB, Claiborne A (June 1990). "Heterogeneity among the flavin-containing NADH peroxidases of group D streptococci. Analysis of the enzyme from Streptococcus faecalis ATCC 9790". J. Biol. Chem. 265 (17): 9857–63. PMID 2161844.
- 1 2 Stehle T, Claiborne A, Schulz GE (January 1993). "NADH binding site and catalysis of NADH peroxidase". Eur. J. Biochem. 211 (1–2): 221–6. doi:10.1111/j.1432-1033.1993.tb19889.x. PMID 8425532.
- 1 2 Yeh JI, Claiborne A (2002). "Crystal structures of oxidized and reduced forms of NADH peroxidase". Meth. Enzymol. Methods in Enzymology. 353: 44–54. doi:10.1016/S0076-6879(02)53035-4. ISBN 978-0-12-182256-9. PMID 12078517.
- ↑ Crane EJ, Yeh JI, Luba J, Claiborne A (August 2000). "Analysis of the kinetic and redox properties of the NADH peroxidase R303M mutant: correlation with the crystal structure". Biochemistry. 39 (34): 10353–64. doi:10.1021/bi000553m. PMID 10956025.
- ↑ Crane EJ, Parsonage D, Poole LB, Claiborne A (October 1995). "Analysis of the kinetic mechanism of enterococcal NADH peroxidase reveals catalytic roles for NADH complexes with both oxidized and two-electron-reduced enzyme forms". Biochemistry. 34 (43): 14114–24. doi:10.1021/bi00043a016. PMID 7578008.
- ↑ Crane EJ, Parsonage D, Claiborne A (February 1996). "The active-site histidine-10 of enterococcal NADH peroxidase is not essential for catalytic activity". Biochemistry. 35 (7): 2380–7. doi:10.1021/bi952347y. PMID 8652580.
- ↑ Dolin MI (March 1957). "The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of a flavin peroxidase for reduced diphosphopyridine nucleotide". J. Biol. Chem. 225 (1): 557–73. PMID 13416259.
- ↑ Dolin MI (September 1977). "DPNH peroxidase: effector activities of DPN". Biochem. Biophys. Res. Commun. 78 (1): 393–400. doi:10.1016/0006-291X(77)91267-0. PMID 199166.
- ↑ Hansson L, Häggström MH (1984). "Effects of growth conditions on the activities of superoxide dismutase and NADH-oxidase/NADH-peroxidase inStreptococcus lactis". Current Microbiology. 10 (6): 345–351. doi:10.1007/BF01626563.
- ↑ Gordon J, Holman RA, McLeod JW (October 1953). "Further observations on the production of hydrogen peroxide by anaerobic bacteria". J Pathol Bacteriol. 66 (2): 527–37. doi:10.1002/path.1700660224. PMID 13118459.
- ↑ Šimonovičová M, Tamás L, Huttová J, Mistrík I (2004). "Effect of Aluminium on Oxidative Stress Related Enzymes Activities in Barley Roots". Biologia Plantarum. 48 (2): 261–266. doi:10.1023/B:BIOP.0000033454.95515.8a.
- ↑ Chen SX, Schopfer P (March 1999). "Hydroxyl-radical production in physiological reactions. A novel function of peroxidase". Eur. J. Biochem. 260 (3): 726–35. doi:10.1046/j.1432-1327.1999.00199.x. PMID 10103001.