BioBIKE
| Initial release | 2002 |
|---|---|
| Development status | Active |
| Written in | Lisp |
| Operating system | Unix-like |
| Available in | English |
| Type | Scientific workflow, Symbolic Computing, Bioinformatics, Artificial Intelligence |
| License | MIT Open Source |
| Website |
BioBIKE |
BioBike[1][2](nee. BioLingua [3]) is a cloud-based, through-the-web programmable (Paas) symbolic biocomputing and bioinformatics platform that aims to make computational biology, and especially intelligent biocomputing (that is, the application of Artificial Intelligence to computational biology) accessible to research scientists who are not expert programmers.[4]
Unique capabilities
BioBIKE is an integrated symbolic biocomputing and bioinformatics platform, built from the start as an entirely (what is now called) cloud-based architecture where all computing is done in remote servers, and all user access is accomplished through web browsers.
BioBIKE has a built-in frame system in which all objects, data, and knowledge are represented. This enables code written either in the native Lisp, in the visual programming language, or systems of rules expressed in the SNARK theorem prover to access the whole of biological knowledge in an integrated manner.
For its time (released in 2002) it was unique in permitting users to create fully functional biocomputing programs that run on the back-end servers entirely through the web browser UI. (In modern terms it was one of the first PaaS (Platform as a Service) systems, predating even Salesforce in this capability.) Initially this programming was carried out in raw Lisp, but Jeff Elhai's team at VCU, with NSF funding, created an entirely graphical programming environment on top of BioBIKE based upon the Boxer-style programming environments.[5]
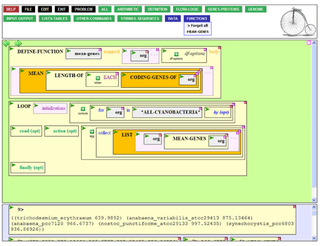
Being a multi-headed, multi-threaded, multi-user, multi-tenancy cloud-based system, BioBIKE users were able to directly work together through their web browsers, remotely sharing the same listerner and memory space. This permitted a unique sort of collaboration, discussed in Shrager (2007).[6]
A specialized offshoot of BioBIKE called "BioDeducta" includes SRI's SNARK theorem prover, offering unique "deductive biocomputing" capabilities.[7]
Implementation
BioBIKE is open-source software implemented using the Lisp programming language. Continuing development takes place by the BioBIKE team[8] centered at Virginia Commonwealth University .
History
BioBIKE was originally called "BioLingua", and was developed by Jeff Shrager at The Carnegie Inst. of Washington Dept. of Plant Biology, and JP Massar with funding from NASA's Astrobiology Division. Shrager and Massar wanted to create a web-based, multi-user Lisp Machine, specialized for bioinformatics. Other early contributors to the project included Mike Travers, and Jeff Elhai of VCU. Elhai obtained continuing funding from the National Science Foundation for the project, which was renamed BioBIKE. Elhai and colleagues added BioBIKE's unique visual programming language. Shrager, meanwhile, collaborated with Richard Waldinger at SRI to build SRI's (SNARK) theorem prover into BioBIKE, creating a deductive biocomputing system, called BioDeducta.[9]

Instances
There are a number of BioBIKE verticals in different biological domains.
See the most current list by clicking here.
See also
External links
References
- ↑ Elhai, J.; Taton, A.; Massar, J.; Myers, J. K.; Travers, M.; Casey, J.; Slupesky, M.; Shrager, J. (2009). "BioBIKE: A Web-based, programmable, integrated biological knowledge base". Nucleic Acids Research. 37 (Web Server issue): W28–W32. doi:10.1093/nar/gkp354. PMC 2703918
 . PMID 19433511.
. PMID 19433511. - ↑ Shrager, J.; Waldinger, R.; Stickel, M.; Massar, J. P. (2007). Futrelle, Robert, ed. "Deductive Biocomputing". PLoS ONE. 2 (4): e339. doi:10.1371/journal.pone.0000339. PMC 1838522
 . PMID 17415407.
. PMID 17415407. - ↑ Massar, J. P.; Travers, M.; Elhai, J.; Shrager, J. (2004). "BioLingua: A programmable knowledge environment for biologists". Bioinformatics. 21 (2): 199–207. doi:10.1093/bioinformatics/bth465. PMID 15308539.
- ↑ Jeff Elhai: Humans, Computers, and the Route to Biological Insights: Regaining Our Capacity for Surprise. Journal of Computational Biology 18(7): 867–878 (2011)
- ↑ Elhai, J.; Taton, A.; Massar, J.; Myers, J. K.; Travers, M.; Casey, J.; Slupesky, M.; Shrager, J. (2009). "BioBIKE: A Web-based, programmable, integrated biological knowledge base". Nucleic Acids Research. 37 (Web Server issue): W28–W32. doi:10.1093/nar/gkp354. PMC 2703918
 . PMID 19433511.
. PMID 19433511. - ↑ J Shrager (2007) The Evolution of BioBike: Community Adaptation of a Biocomputing Platform. Studies in History and Philosophy of Science, 38, 642–656.
- ↑ Shrager, J.; Waldinger, R.; Stickel, M.; Massar, J. P. (2007). Futrelle, Robert, ed. "Deductive Biocomputing". PLoS ONE. 2 (4): e339. doi:10.1371/journal.pone.0000339. PMC 1838522
 . PMID 17415407.
. PMID 17415407. - ↑ http://biobike.org/
- ↑ Shrager, J.; Waldinger, R.; Stickel, M.; Massar, J. P. (2007). Futrelle, Robert, ed. "Deductive Biocomputing". PLoS ONE. 2 (4): e339. doi:10.1371/journal.pone.0000339. PMC 1838522
 . PMID 17415407.
. PMID 17415407.