Beta turn
Beta turns (β turns, β-turns, β-bends, tight turns, reverse turns) are very common motifs in proteins and polypeptides.[1][2][3][4][5][6][7][8][9] Each consists of four amino acid residues (labelled i, i+1, i+2 and i+3). They can be defined in two ways: 1. By the possession of an intra-main-chain hydrogen bond between the CO of residue i and the NH of residue i+3. Alternatively, 2. By having a distance of less than 7Å between the Cα atoms of residues i and i+3. The hydrogen bond criterion is the one most appropriate for everyday use, partly because it gives rise to four distinct categories; the distance criterion gives rise to the same four categories but yields additional turn types. Two websites are available for finding and examining hydrogen-bonded beta turns in proteins, Motivated Proteins: [10] and PDBeMotif .[11]
Beta turns defined by hydrogen bond criterion:
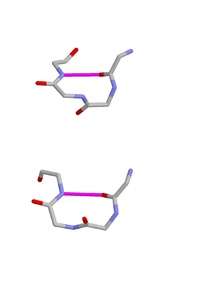
The hydrogen bond criterion for beta turns, applied to polypeptides whose amino acids are linked by trans peptide bonds, gives rise to just four categories, as shown by Venkatachalam in 1968. They are called types I, II, I’ and II’. All occur regularly in proteins and polypeptides but type I is commonest, because it most resembles an alpha helix, occurring within 3/10 helices and at the ends of some classic alpha helices.
The four types of beta turn are distinguished by the Φ,ψ angles of residues i+1 and i+2 as shown in the Table below giving the typical average values. Glycines are especially common at amino acids with positive Φ angles; for prolines such a conformation is sterically impossible but they occur frequently at amino acid positions where Φ is negative.
| Φi+1 | ψi+1 | Φi+2 | ψi+2 | |
|---|---|---|---|---|
| type I | -60 | -30 | -90 | 0 |
| type II | -60 | 120 | 80 | 0 |
| type I' | 60 | 30 | 90 | 0 |
| type II' | 60 | -120 | -80 | 0 |
The main chain atoms of type I and I’ β turns are enantiomers (mirror images) of one another. So are the main chain atoms of type II and II’ β turns.
Type I and II β turns exhibit a relationship to one another because they potentially interconvert by the process of peptide plane flipping (180° rotation of the CONH peptide plane with little positional alteration to side chains and surrounding peptides). The same relationship exists between type I’ and II’ β turns. Some evidence has indicated that these interconversions occur in beta turns in proteins such that crystal or NMR structures merely provide a snapshot of β turns that are, in reality, interchanging.[12] In proteins in general all four beta turn types occur frequently but I is commonest, followed by II, I' and II' in that order. Beta turns are especially common at the loop ends of beta hairpins; they have a different distribution of types from the others; type I' is commonest, followed by types II', I and II.
Asx turns and ST turns resemble beta turns except that residue i is replaced by the side chain of an aspartate, asparagine, serine or threonine. The main chain - main chain hydrogen bond is replaced by a side chain – main chain hydrogen bond. 3D computer superimposition shows that, in proteins, they occur [13] as one of the same four types that beta turns do except that their relative frequency of occurrence differs: Type II’ is commonest, followed by types I, II and I’.
Beta turns defined by distance criterion:
Apart from the type I, I’,II and II’ beta turns as identified via the hydrogen bond criterion, non-hydrogen-bonded beta-turns named type VIII often occur. Three other, fairly rare, types of beta turn have been identified in which the peptide bond between residues i+1 and i+2 is cis rather than trans; these are named types VIa1, VIa2 and VIb. Another category, type IV, was used for turns not belonging to any of the above. Further details of these turns are given in turn (biochemistry).
References
- ↑ Venkatachalam, CM (1968). "Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units". Biopolymers. 6 (10): 1425–1436. doi:10.1002/bip.1968.360061006. PMID 5685102.
- ↑ Lewis, PN; Momany FA (1973). "Chain reversal in proteins". Biochim Biophys Acta. 303 (2): 211–209. doi:10.1016/0005-2795(73)90350-4. PMID 4351002.
- ↑ Toniolo, C; Benedetti E (1980). "Intramolecularly hydrogen-bonded peptide conformations". CRC Crit Rev Biochem. 9 (1): 1–44. doi:10.3109/10409238009105471. PMID 6254725.
- ↑ Richardson, JS (1981). "The anatomy and taxonomy of protein structure". Adv Prot Chem. 34: 167–339. doi:10.1016/S0065-3233(08)60520-3. PMID 7020376.
- ↑ Rose, GD; Gierasch LM (1985). "Turns in peptides and proteins". Adv Prot Chem. 37: 1–109. doi:10.1016/S0065-3233(08)60063-7. PMID 2865874.
- ↑ Milner-White, EJ; Poet R (1987). "Loops, bulges, turns and hairpins in proteins". Trends Biochem Sci. 12: 189–192. doi:10.1016/0968-0004(87)90091-0.
- ↑ Wilmot, CM; Thornton JM (1988). "Analysis and prediction of the different types of beta-turn in proteins". J Mol Biol. 203: 221–232. doi:10.1016/0022-2836(88)90103-9. PMID 3184187.
- ↑ Sibanda, BL; Blundell TL (1989). "Conformation of β-hairpins in protein structures: A systematic classification with applications to modelling by homology, electron density fitting and protein engineering". J Mol Biol. 206 (4): 759–777. doi:10.1016/0022-2836(89)90583-4. PMID 2500530.
- ↑ Hutchinson, EG; Thornton JM (1994). "A revised set of potentials for β-turn formation in proteins". J Mol Biol. 3 (12): 2207–2216. doi:10.1002/pro.5560031206. PMC 2142776
 . PMID 7756980.
. PMID 7756980. - ↑ Leader, DP; Milner-White EJ (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs.". BMC Bioinformatics. 10 (1): 60. doi:10.1186/1471-2105-10-60. PMC 2651126
 . PMID 19210785.
. PMID 19210785. - ↑ Golovin, A; Henrick K (2008). "MSDmotif: exploring protein sites and motifs.". BMC Bioinformatics. 9 (1): 312. doi:10.1186/1471-2105-9-312. PMC 2491636
 . PMID 18637174.
. PMID 18637174. - ↑ Hayward, S (2001). "Peptide plane flipping in proteins". Protein Science. 10 (11): 2219–2227. doi:10.1110/ps.23101. PMC 2374056
 . PMID 11604529.
. PMID 11604529. - ↑ Duddy, WM; Nissink JWM (2004). "Mimicry by asx- and ST-turns of the four main types of β turn in proteins". Protein Science. 13 (11): 3051–3055. doi:10.1110/ps.04920904. PMID 15459339.